New avenues in fight against drug resistant TB
While rising drug resistance in Mycobacterium tuberculosis is eroding the power of almost all available antibiotics in the fight against this major pathogen, work completed by MWC-supported postgraduate students and emerging researchers is unveiling new promising and complementary areas of attack targeting tuberculosis (TB) and antibiotic resistance.
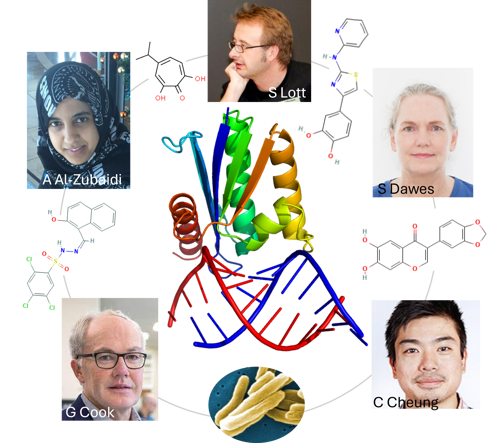
MWC team members Dr Al-Zubaidi, Dr Dawes and A/P Lott (University of Auckland) and Prof Cook, Mr Cheung (University of Otago ) validated RNase HI, an enzyme that destroys RNA:DNA hybrids, as a new vulnerable drug target in Mycobacterium tuberculosis (image insert at the bottom). They discovered that inhibition of RNase HI makes rifampicin more potent, which might help redesign first-line therapy for tuberculosis, and limit antimicrobial resistance. Outer circle: Chemical structures of the four RNase HI inhibitor compounds that potentiate killing of M. tuberculosis by rifampicin. Centre: Cartoon drawing of RNase HI (top) in complex with RNA:DNA hybrid (bottom). Image courtesy of Stephanie Dawes.
Multidrug-resistant TB is defined by the resistance of M. tuberculosis to first-line antibiotics rifampicin and isoniazid. Mitigating or reversing resistance to these inexpensive first-line drugs offers a means of preserving and extending their use in TB treatment.
With support from the MWC, Dr Abeer Al-Zubaidi, alongside other MWC researchers led by Dr Stephanie Dawes and Associate Professor Shaun Lott, have discovered a new means of enhancing the potency of rifampicin and other anti-TB drugs. The University of Auckland team has also identified promising compounds providing a first step in the development of a new class of antimycobacterial drug.
Parallel work by University of Otago PhD candidate Natalie Waller and senior researcher Dr Matthew McNeil has uncovered weaknesses in drug resistant strains of M. tuberculosis that if exploited could lead to new unique drug combinations to greatly reduce treatment times from months to weeks and prevent the emergence of new drug resistance.
Work from both teams has been published in two high impact journals Antimicrobial Agents and Chemotherapy1, and Nature Communications2.
Stephanie and Shaun’s team set their focus on R-loop metabolism. R loops can be lethal to the cell if not resolved. RNase HI is an enzyme that removes R-loops. By depleting the RNase HI enzyme in mycobacteria, sensitivity to Rifampicin increased by almost 100-fold.
Stephanie points to the potential of an RNase HI inhibitor which could be used in conjunction with Rifampicin.
“Rifampicin has many side effects. An RNase HI inhibitor could enhance first-line antibiotic activity. Patients could receive the same benefits from a fraction of a dose, and in turn have fewer side effects and greater likelihood of completing the course of drugs. A good RNase HI inhibitor may also re-sensitise rifampicin-resistant bacteria, enabling the drug to be used again on resistant strains.”
Researchers also identified four small molecules known to inhibit RNase HI from HIV which could be re-purposed to inhibit M. tuberculosis.
Looking at the entirely different phenomenon of collateral sensitivity, work done by MWC researchers Natalie Waller and Matthew McNeil found new ways to kill drug-resistant strains of M. tuberculosis and new ways to combine different drugs to stop drug resistance from occurring in the first place.
When a pathogen becomes resistant to a specific antibiotic, it can become vulnerable or sensitive to another unrelated antibiotic. By identifying this susceptibility by drug susceptibility profiling, genomics and evolutionary studies, the MWC researchers show drug resistance strains could be killed off rapidly.
McNeil says “this work, in combination with the contributions of others, highlights how an improved understanding of the biology and consequences of drug-resistance can identify unique therapeutic strategies to revolutionize the fight against anti-microbial resistance.”
The discovery has promising implications for other drug-resistant diseases.